Transactions on Cryptographic Hardware and Embedded Systems 2025
Adaptive Template Attacks on the Kyber Binomial Sampler
README
Adaptive Template Attacks on the Kyber Binomial Sampler
Eric Chun-Yu Peng, Markus G. Kuhn
This repository contains the Julia code needed for reproducing the experiments described in our paper
Eric Chun-Yu Peng, Markus G. Kuhn: Adaptive Template Attacks on the Kyber Binomial Sampler. IACR Transactions on Cryptographic Hardware and Embedded Systems, Vol. 2025, No. 3.
Requirements
This code was mainly developed and tested on x86-64 (64-bit) computers running Ubuntu Linux 20.04 or 24.04 with 48 GB RAM. We hope it will run on any platform that is supported by Julia and has at least 16 GB RAM. About 25 GB disk space would be ideal, but there are also ways to run the demo with only about 12 GB disk space.
Install Julia
To run this code, you will need Julia (version 1.11 or newer).
For Linux or macOS, best install Julia via the juliaup installation manager using the following shell command line:
$ curl -fsSL https://install.julialang.org | sh
Download this repository
Clone this repository and its submodules to your local machine, then install (“instantiate”) the required Julia package dependencies for this project into your Julia depot path (default: ~/.julia/):
$ git clone --recursive https://github.com/eric-cyp24/Kyber768cbd.jl.git
$ cd Kyber768cbd.jl
$ julia --project -e "import Pkg; Pkg.instantiate()"
Download trace data for DK2 and MS2
This repository contains only Julia code. To reproduce some of the experiments from the paper, you also need to separately download the data used.
The following Julia script will download preprocessed traces for two devices (4000 traces to profile DK2 and 1000 traces each to attack KeyGen (K) and Encaps (E) on MS2). It will also download pre-built post-LDA templates for the other devices.
(The downloaded traces have already been downsampled from 2.5 GHz to 100 MHz using a Lanczos anti-aliasing filter, and have then been cut into 48 segments each, as detailed in sections 3, 4.3 and 5.3 of the paper.)
The script will download this data into the folder data/ (into subfolder Traces/), unless you specify another folder using the environment variable DATA_DIR. This folder will later also be used for temporary data.
Choose one of the following options, depending on how much disk space you have:
Option 1: if you have the space, download all provided data at once with
$ julia --project scripts/downloaddata.jl # requires 19 GBOption 2: download data needed for profiling and attack separately, as indicated below, using the options
$ julia --project scripts/downloaddata.jl --profiling # requires 8.1 GB $ julia --project scripts/downloaddata.jl --attack # requires 4.9 GBright before the profiling and attack steps. You can then remove them immediately after the corresponding processing step, to reduce the required disk space, using
$ julia --project scripts/deletedata.jl --profiling # frees 8.1 GB $ julia --project scripts/deletedata.jl --attack # frees 4.9 GBrespectively.
The above allows you to reproduce all the attacks against the MS2 device, as well as the profiling steps for the DK2 device.
Optional: You can also download the profiling and attack outcomes, including all the LDA projections and adjusted template parameters for all the other profiling and target device combinations:
$ julia --project scripts/downloaddata.jl --results # requires 6.0 GB
Build tables 2 and 3
You can run the following Julia script to generate the LaTeX table source code for tables 2 and 3, and pdflatex to generate these tables as results/SuccessRateTables.pdf.
$ julia --project scripts/h5result2latextable_multiboardsingletrace.jl
$ pdflatex -output-directory results scripts/SuccessRateTables.tex
If you downloaded the results for other target devices than MS2, you should see those columns already completed:
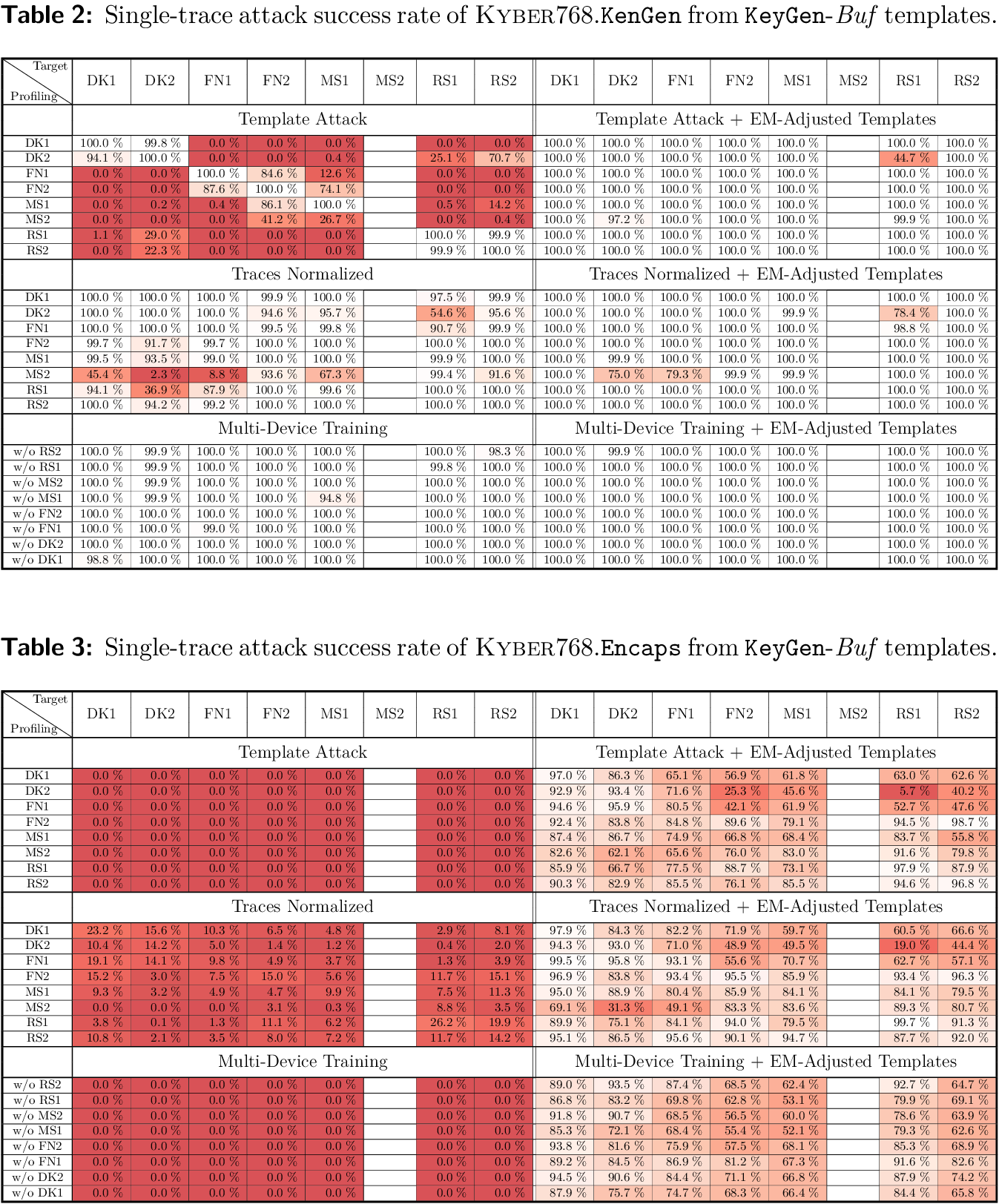
To complete the still missing MS2 columns:
- First, build the DK2 profiling templates with the following profiling step.
- Then, run the single-trace attacks on the MS2 target with templates from all devices (DK1–RS2).
Profiling
Build the LDA-based templates from profiling traces of device DK2: data/Traces/SOCKET_HPF/DK2/test_20241219/lanczos2_25/traces_lanczos2_25_proc.h5. The resulting templates will be stored in the folder data/Traces/SOCKET_HPF/DK2/test_20241219/lanczos2_25/Templates_POIe40-80/.
$ julia --project scripts/downloaddata.jl --profiling # for Option 2 only
$ julia --project scripts/profiling_kyber768cbd.jl
$ julia --project scripts/deletedata.jl --profiling # for Option 2 only
Single-trace attacks
Run the single-trace attacks on the MS2 target device:
$ julia --project scripts/downloaddata.jl --attack # for Option 2 only
$ julia --project scripts/attack_kyber768cbd_Buf_singletrace.jl
You can speed up the attack by running Julia with multiple threads:
$ julia --project -t4 scripts/attack_kyber768cbd_Buf_singletrace.jl
Option -t4 tells Julia to execute with 4 threads. Adjust this based on your computer's resources, i.e. speed up the computation with more threads or decrease RAM usage with fewer. Each attack trace set (table cell) takes 3–15 minutes to finish. Therefore, this script may run for a while (roughly 4–5 hours on our computers).
The attack results will be stored as HDF5 files in folder data/Traces/SOCKET_HPF/MS2/test_20241221/lanczos2_25_test_K/Results/Templates_POIe40-80/.
To see more detailed attack metadata, use an HDF5 viewer, e.g. HDFView Software or myHDF5 online viewer; or open the file directly with the HDF5.jl package in Julia, as in
$ julia --project
julia> using HDF5
julia> h = h5open("data/Traces/SOCKET_HPF/MS2/test_20241221/lanczos2_25_test_K/Results/Templates_POIe40-80/marginalize_Buf_Result_with_Templates_POIe40-80_from_SOCKET_HPF_DK2_test_20241219.h5")
julia> h["Traces_Normalized_Templates_Adj_EM"]["success_rate_single_trace"][]
0.777
Attacking the Encaps operation
To attack the Encaps operation (Table 3 above), run the same attack script with the additional command line argument: --targetOP Encaps
$ julia --project scripts/attack_kyber768cbd_Buf_singletrace.jl --targetOP Encaps
Generate (LaTeX) tables
To view the single-trace attack results, run the following code (again) to generate results/SuccessRateTables.pdf with the newly produced success rates:
$ julia --project scripts/h5result2latextable_multiboardsingletrace.jl
$ pdflatex -output-directory results scripts/SuccessRateTables.tex
Adaptive template attack evaluation
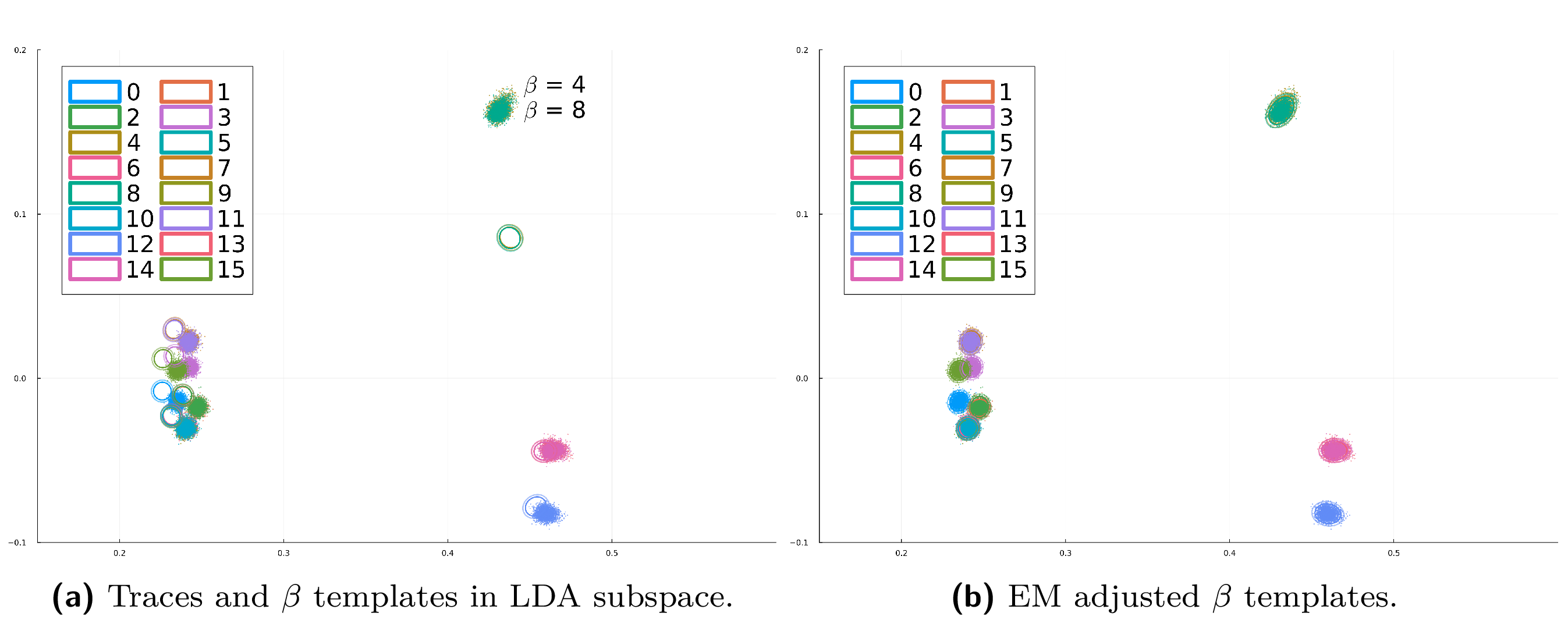
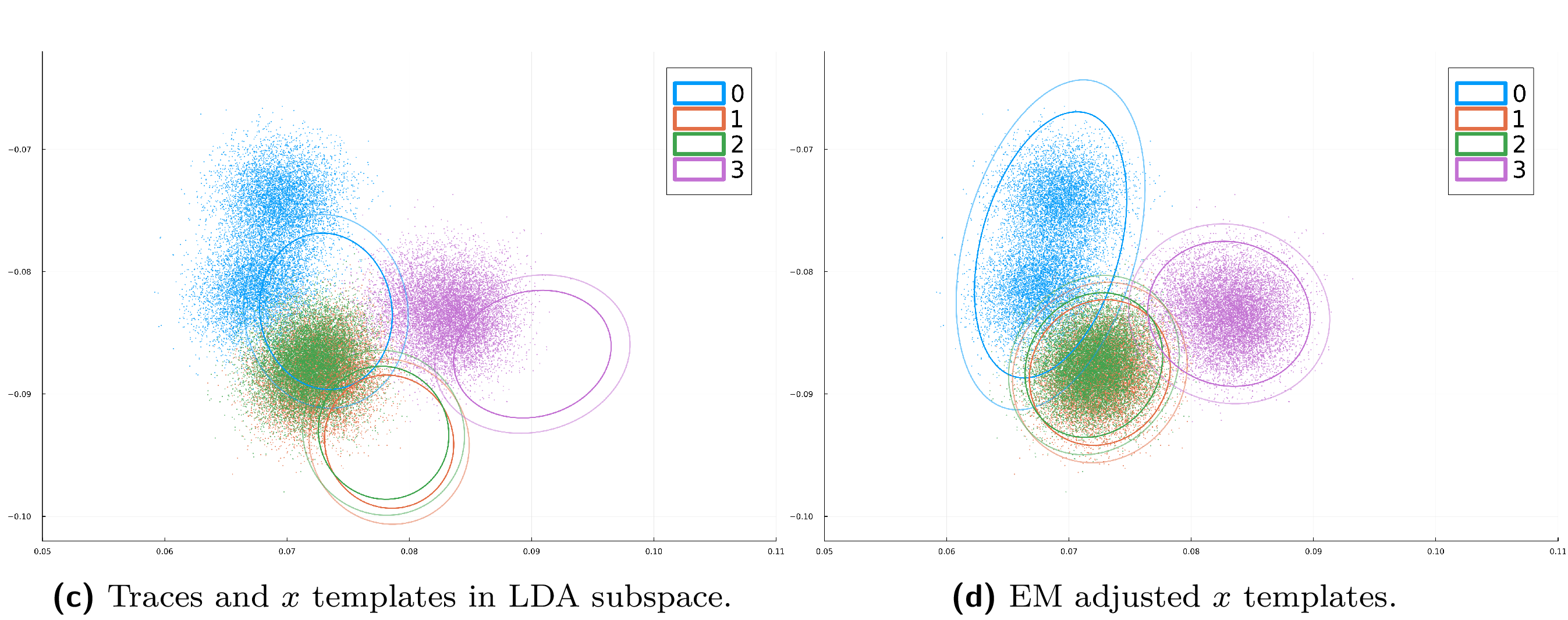
To view the effect of the EM-based template adjustment, run the following script:
$ julia --project scripts/figure_emadj_templates.jl --variable XY --output results/traces_and_XY_templates.png
This generates the before-and-after plots (a) and (b) from Figure 5, stored in the results/ folder as traces_and_XY_templates.png and traces_and_XY_templates_EMadj.png.
If you want to see the effect of EM template adjustment on a lower-leakage intermediate variable (iv), modify the command line options for the script:
$ julia --project scripts/figure_emadj_templates.jl --variable X --output results/traces_and_X_templates.png
You should now see plots (c) and (d) from Figure 5 in traces_and_X_templates.png and traces_and_X_templates_EMadj.png, also in the results/ folder, similar to the ones shown below.